- Lab Credits: Dhanush Kumar Shankar
- Blog Credits: Sai Akhilesh Ande
Note:
- Currenlty Airflow (
pip install apache-airflow) installation is only supported for Linux based systems(MacOS, Ubuntu, etc.). On Windows, you can install it via WSL2 or use (virtual box/vmware) to install a Linux OS. - However, this specific lab uses Airflow’s Docker image to run Airflow (without any installation). Hence, this specific lab can be run on Windows, MacOS and Linux.
- It is important to understand that this lab doesn’t require any virtual environment or installation of packages. We will just just specify all required packages to Docker and it will install them in a container environment. That’s the advantage of Docker.
This lab revolves around two key modules:
- Docker: A platform for developing, shipping, and running applications in containers.
- Airflow: A platform to programmatically author, schedule, and monitor workflows.
In this lab, we create a machine-learning workflow using Airflow which automates a task of data clustering using K-Means clustering and determining optimal number of clusters with the elbow method.
The workflow involves the following steps:
- Load data from a CSV file.
- Perform data preprocessing.
- Build and save a K-Means clustering model.
- Load the saved model and determine the optimal number of clusters based on the elbow method.
Prerequisites
- Github Lab-1
- Docker Desktop Installed
- Basics of Docker - containers and images.
Setting up the lab
Note: If you’d like to save your work files to GitHub, setup your working directory similar to Github Lab-1 and add necessary files to .gitignore. If you just want to run the lab locally, follow the below steps.
- Open the local mlops_labs directory using Visual Studio Code(VSC) and open a new terminal(use cmd for windows) inside VSC.
- Create a new working directory for this lab(e.g. airflow_lab1).
- Move into the working directory using the command cd airflow_lab1 in your terminal.
Getting the necessary files
Get the required files for this lab from the MLOps repo.
Project Structure:
mlops_labs/
└── airflow_lab1/
└── dags/
├── data/
│ ├── file.csv
│ └── test.csv
├── model/
├── src/
│ ├── __init__.py
│ └── lab.py
└── airflow.py
Airflow
We use Airflow to author workflows as directed acyclic graphs (DAGs) of tasks. The Airflow scheduler executes your tasks on an array of workers while following the specified dependencies.
References:
Prerequisites:
- Local
- Docker Desktop Running
- Cloud
- Linux VM
- SSH Connection
- Installed Docker Engine - Install using the convenience script
You should allocate at least 4GB memory for the Docker Engine (ideally 8GB).
Running an Apache Airflow DAG Pipeline in Docker
This guide provides detailed steps to set up and run an Apache Airflow Directed Acyclic Graph (DAG) pipeline within a Docker container using Docker Compose. The pipeline is named Airflow_Lab1”.
- Make sure
Docker Desktopis up and running. - In this lab, we will run
AirflowinsideDocker. Hence, this lab doesn’t have arequirements.txtfile. - You can check if you have enough memory by running this command (optional).
docker run --rm "debian:bullseye-slim" bash -c 'numfmt --to iec $(echo $(($(getconf _PHYS_PAGES) * $(getconf PAGE_SIZE))))' - Fetch
docker-compose.yaml.# mac and linux users curl -LfO 'https://airflow.apache.org/docs/apache-airflow/2.9.2/docker-compose.yaml' # windows users curl -o docker-compose.yaml https://airflow.apache.org/docs/apache-airflow/2.9.2/docker-compose.yaml - Create required directories.
# mac and linux users mkdir -p ./dags ./logs ./plugins ./config # windows users (if using cmd) mkdir dags logs plugins config # windows users (if using powershell) mkdir dags, logs, plugins, config - Setting the right Airflow user. This will create a
.envfile.echo -e "AIRFLOW_UID=$(id -u)" > .env- If you are on windows platform, replace the content of
.envfile with the following.AIRFLOW_UID=50000
- If you are on windows platform, replace the content of
- Update the following in
docker-compose.yml.# Donot load examples AIRFLOW__CORE__LOAD_EXAMPLES: 'false' # Additional python packages _PIP_ADDITIONAL_REQUIREMENTS: ${_PIP_ADDITIONAL_REQUIREMENTS:- pandas scikit-learn kneed} # Output dir - ${AIRFLOW_PROJ_DIR:-.}/working_data:/opt/airflow/working_data # Change default admin credentials _AIRFLOW_WWW_USER_USERNAME: ${_AIRFLOW_WWW_USER_USERNAME:-airflow2} _AIRFLOW_WWW_USER_PASSWORD: ${_AIRFLOW_WWW_USER_PASSWORD:-airflow2} - Initialize the database. This will take a couple of minutes.
docker compose up airflow-init - Run Airflow.
docker compose upWait until terminal outputs
airflow-webserver-1 | 127.0.0.1 - - [19/Feb/2024:17:16:53 +0000] "GET /health HTTP/1.1" 200 318 "-" "curl/7.88.1" - Enable port forwarding. Windows users click “allow” in the pop-up window.
- Visit
localhost:8080and login with credentials set on step2.d. - You will see your dag(Airflow_Lab1) as shown in the below image. For this lab, you are already given a dag script -
dags/airflow.py. You can create more dags with similar scripts.
You can create more dags with similar scripts.
- BashOperator
- PythonOperator
- Task Dependencies
- Params
- Crontab schedules
You can have n number of scripts inside the
dagsdirectory. - To manually trigger the DAG, click on the “Trigger DAG” button or enable the DAG by toggling the switch to the “On” position.
- Monitor the progress of the DAG in the Airflow web interface. You can view logs, task status, and task execution details in airflow UI and also in
logsdirectory. - Once the DAG completes its execution, check any output or artifacts produced by your functions and tasks.
- Monitor the progress of the DAG in the Airflow web interface. You can view logs, task status, and task execution details in airflow UI and also in
- Click on your dag(Airflow_Lab1) -> Graph tab -> click on the last rectangle box in the graph(load_model_task) -> Logs tab. You will find the result of the workflow i.e., optimal number of clusters as shown in the below image.
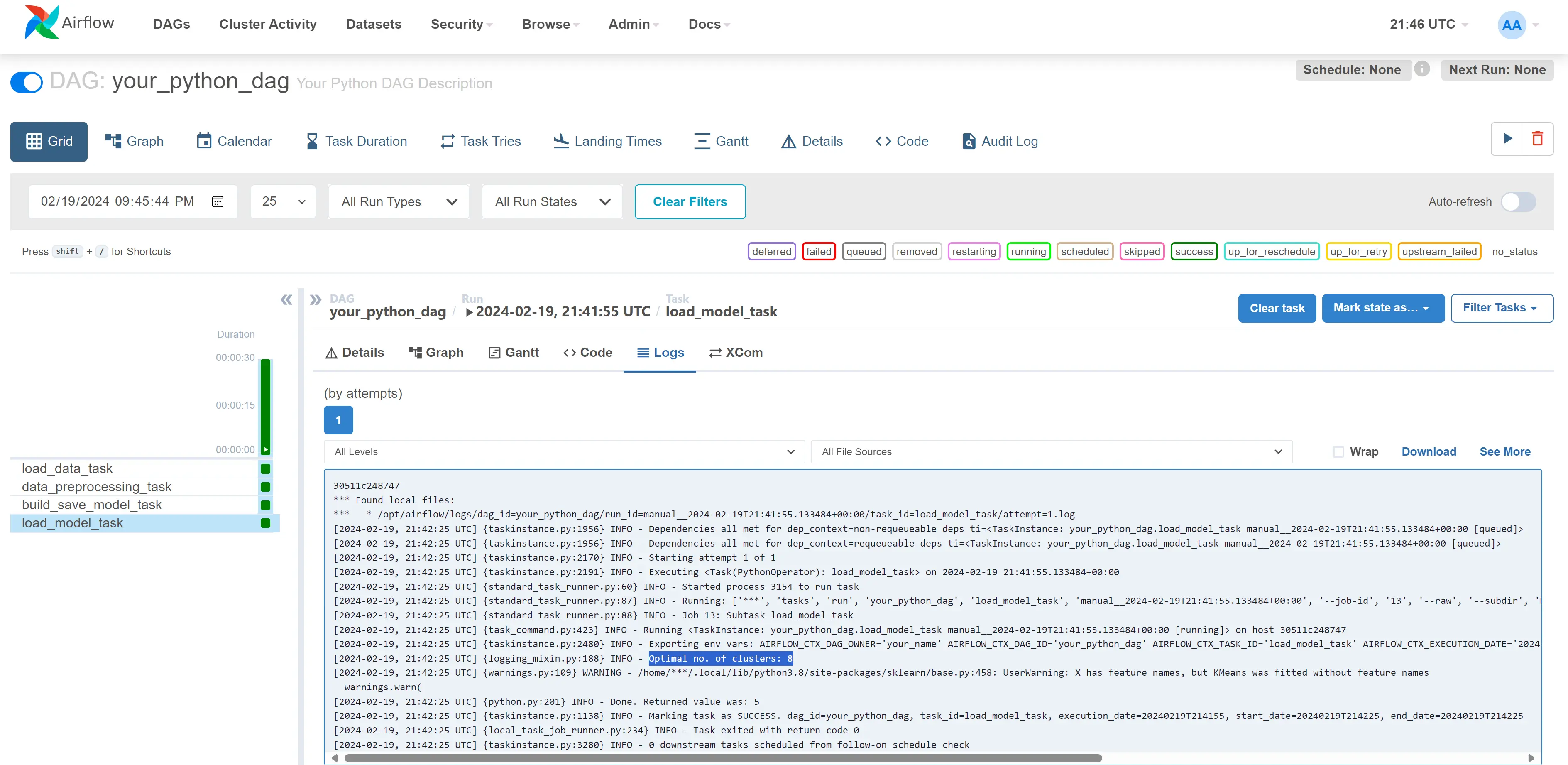
- Explore UI and add user
Security > List Users. (optional) - To stop Airflow, we have to stop the docker containers. For this, we have to open a new terminal(cmd for windows), activate the virtual enviroment and run the following command.
docker compose down
Final Project Structure:
mlops_labs/
└── airflow_lab1/
├── config/
├── dags/
│ ├── data/
│ │ ├── file.csv
│ │ └── test.csv
│ ├── model/
│ │ └── model.sav
│ ├── src/
│ │ ├── __init__.py
│ │ └── lab.py #
│ └── airflow.py # Your DAG script
├── logs/
├── plugins/
├── .env
└── docker-compose.yaml
Note: If your working in a git repo, make sure to add __pycache__ to .gitignore.
Airflow DAG Script
This part of the lab provides a detailed explanation of the dags/airflow.py python script that defines an Airflow Directed Acyclic Graph (DAG) for a data processing and modeling workflow.
Script Overview
The script defines an Airflow DAG named Airflow_Lab1 that consists of several tasks. Each task represents a specific operation in a data processing and modeling workflow. The script imports necessary libraries, sets default arguments for the DAG, creates PythonOperators for each task, defines task dependencies, and provides command-line interaction with the DAG.
Importing Libraries
# Import necessary libraries and modules
from airflow import DAG
from airflow.operators.python_operator import PythonOperator
from datetime import datetime, timedelta
from src.lab import load_data, data_preprocessing, build_save_model, load_model_elbow
from airflow import configuration as conf
The script starts by importing the required libraries and modules. Notable imports include the DAG and PythonOperator classes from the airflow package, datetime manipulation functions, and custom functions from the src.lab module.
Enable pickle support for XCom, allowing data to be passed between tasks
conf.set('core', 'enable_xcom_pickling', 'True')
Define default arguments for your DAG
default_args = {
'owner': 'your_name',
'start_date': datetime(2023, 9, 17),
'retries': 0, # Number of retries in case of task failure
'retry_delay': timedelta(minutes=5), # Delay before retries
}
Default arguments for the DAG are specified in a dictionary named default_args. These arguments include the DAG owner’s name, the start date, the number of retries, and the retry delay in case of task failure.
Create a DAG instance named ‘Airflow_Lab1’ with the defined default arguments
dag = DAG(
'Airflow_Lab1',
default_args=default_args,
description='Your Python DAG Description',
schedule_interval=None, # Set the schedule interval or use None for manual triggering
catchup=False,
)
Here, the DAG object is created with the name Airflow_Lab1 and the specified default arguments. The description provides a brief description of the DAG, and schedule_interval defines the execution schedule (in this case, it’s set to None for manual triggering). catchup is set to False to prevent backfilling of missed runs.
Task to load data, calls the ‘load_data’ Python function
load_data_task = PythonOperator(
task_id='load_data_task',
python_callable=load_data,
dag=dag,
)
Task to perform data preprocessing, depends on ‘load_data_task’
data_preprocessing_task = PythonOperator(
task_id='data_preprocessing_task',
python_callable=data_preprocessing,
op_args=[load_data_task.output],
dag=dag,
)
The data_preprocessing_task depends on the load_data_task and calls the data_preprocessing function, which is provided with the output of the load_data_task`.
Task to build and save a model, depends on ‘data_preprocessing_task’
build_save_model_task = PythonOperator(
task_id='build_save_model_task',
python_callable=build_save_model,
op_args=[data_preprocessing_task.output, "model.sav"],
provide_context=True,
dag=dag,
)
The build_save_model_task depends on the data_preprocessing_task and calls the build_save_model function. It also provides additional context information and arguments.
Task to load a model using the ‘load_model_elbow’ function, depends on ‘build_save_model_task’
load_model_task = PythonOperator(
task_id='load_model_task',
python_callable=load_model_elbow,
op_args=["model.sav", build_save_model_task.output],
dag=dag,
)
The load_model_task depends on the build_save_model_task and calls the load_model_elbow function with specific arguments.
Set task dependencies
load_data_task >> data_preprocessing_task >> build_save_model_task >> load_model_task
The task dependencies are defined using the » operator. In this case, the tasks are executed in sequence: load_data_task -> data_preprocessing_task -> build_save_model_task -> load_model_task.
If this script is run directly, allow command-line interaction with the DAG
if __name__ == "__main__":
dag.cli()
- Lastly, the script allows for command-line interaction with the DAG. When the script is run directly, the dag.cli() function is called, providing the ability to trigger and manage the DAG from the command line.
Usage for your custom dataset
You can modify this script to perform K-Means clustering on your own dataset keeping the high-level functions same as follows:
# Load the data
data = load_data()
# Preprocess the data
preprocessed_data = data_preprocessing(data)
# Build and save the clustering model
sse_values = build_save_model(preprocessed_data, 'clustering_model.pkl')
# Load the saved model and determine the number of clusters
result = load_model_elbow('clustering_model.pkl', sse_values)
print(result)
Functions
- load_data():
- Description: Loads data from a CSV file, serializes it, and returns the serialized data.
- Usage:
data = load_data()
- data_preprocessing(data)
- Description: Deserializes data, performs data preprocessing, and returns serialized clustered data.
- Usage:
preprocessed_data = data_preprocessing(data)
- build_save_model(data, filename)
- Description: Builds a K-Means clustering model, saves it to a file, and returns SSE values.
- Usage:
sse_values = build_save_model(preprocessed_data, 'clustering_model.pkl')
- load_model_elbow(filename, sse)
- Description: Loads a saved K-Means clustering model and determines the number of clusters using the elbow method.
- Usage:
result = load_model_elbow('clustering_model.pkl', sse_values)